How to set up your scientific project on the PC? Here is how I do it – from knowledge management over GitHub to this website.
I love preparing projects – maybe this is the most loved part of every project for me. Thinking about all the crazy stuff you will do in the future, letting thoughts fly around and connecting all kinds of ideas and perspectives together. Beeing creative about practice. But maybe it is all about trying to prevent troubles in the future.
The process described here is always at the beginning, long time before I know what I will do exactly in my research, course or thesis. More specific processes like literature research or how I use iPython for data analysis will follow.
Knowledge management
It always starts with organizing knowledge. Writting every thought together and trying to find a structure for it. Some thoughts start on a paper, some directly in my .org file.
Mostly I seperate them in those sections:
- ToDo for tasks of course. Later on in big projects there are subsections seperated in sequences like now, next week, after publication etc.
- Timeline for general planing and scheduling. Some planing and scheduling for very important process-steps (like research) will be saved in the specific section of it. This decision is always a checks and balance between having central overview and having it where you work on something specific.
- Notes for saving all kinds of thoughts and facts.
- Documentation for the ongoing documentation of my work, like content for my next blog posts as well as schedules for it.
- Research for all I need to save research specific. Questions around the hypothesis, data structure, notes on results and so on, but just for a specific problem.
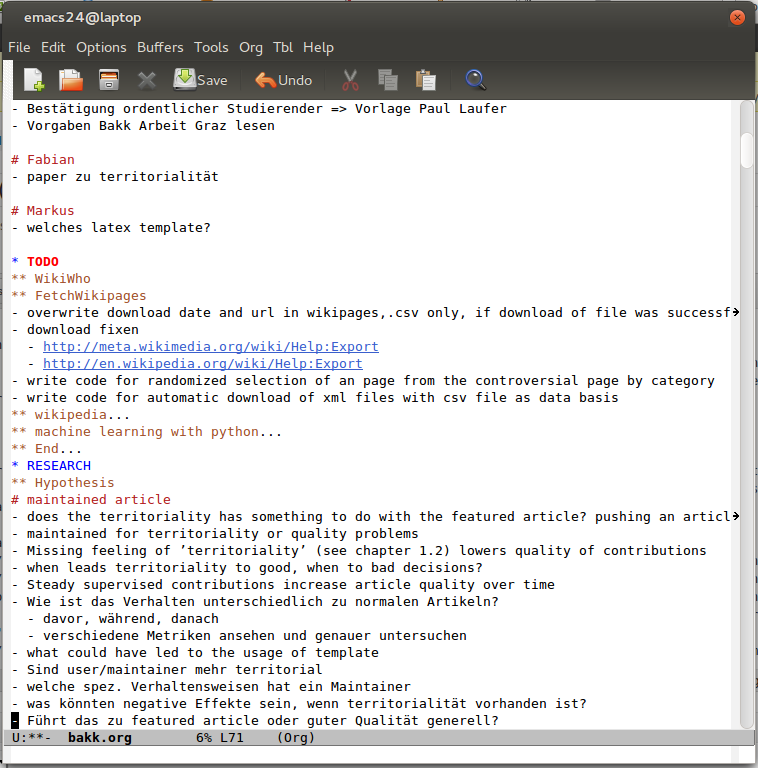
*.org-file for knowledge management
All this is rather flexible and changes from project to project a little bit. But why do I not share my *.org-file on a regular basis? It is sometimes really messy, things for private purpose or with security restrictions are in, or german written notes. Basically, it is very important and always under change and so the effort to keep it always able to be published would be way too many effort for me right now.
GitHub
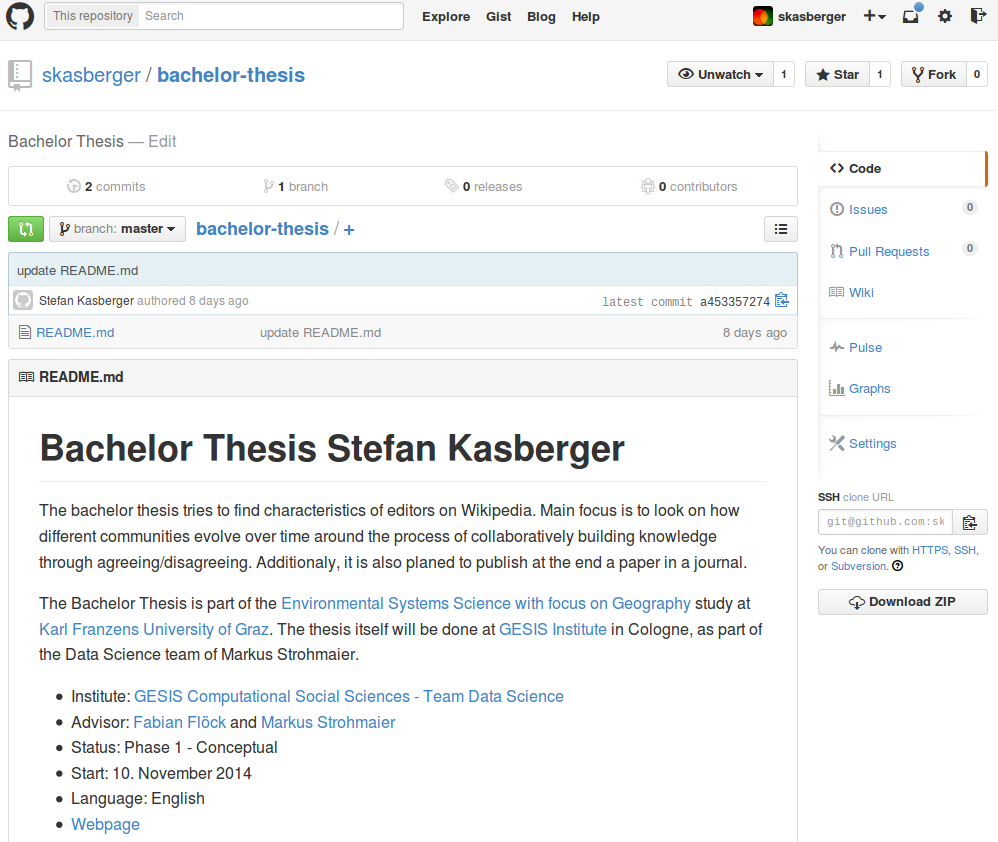
Second step is to create an own folder and initialize in it as git repository for later uploading onto GitHub. As usual, I start with creating the folder structure at my local harddrive. It is not always the same, but seems like some things reoccure.
- applications: Sometimes, applications have their special files, like project files or templates.
- code: One subfolder for the sourcecode of every used programming language, like python, shell, c and so on.
- data: Data collected and created for quantitative analysis. Subfolders are raw (for raw data) and different file-types like json, csv, shape, etc (seperation also inside raw folder). For sharing the data later on, the file-size and file-type is crucial. GitHub is working line based, so it can not create incremental updates of binary files and the file-size is limited to 100mb. So for big datasets I recommend other repositories, like Figshare or domain specific ones.
- docs: In here is all the literature, notes about it and all other related documents before I save it in my Zotero library. Most documents in here will never be published cause of copyright restrictions! I will write an own post about my literature research process with zotero and how my citation and notes archive work.
- images: Content depends on usage, but mostly figures created through the analysis. A raw folder is inside for figures which will not be published afterwards and an final folder for the published ones. Also pictures for website or other documentation purpose can be saved.
- reports: Inside are all reports, mostly written in LaTeX. Every report is one subfolder (except there is only one) with a subfolder for images.
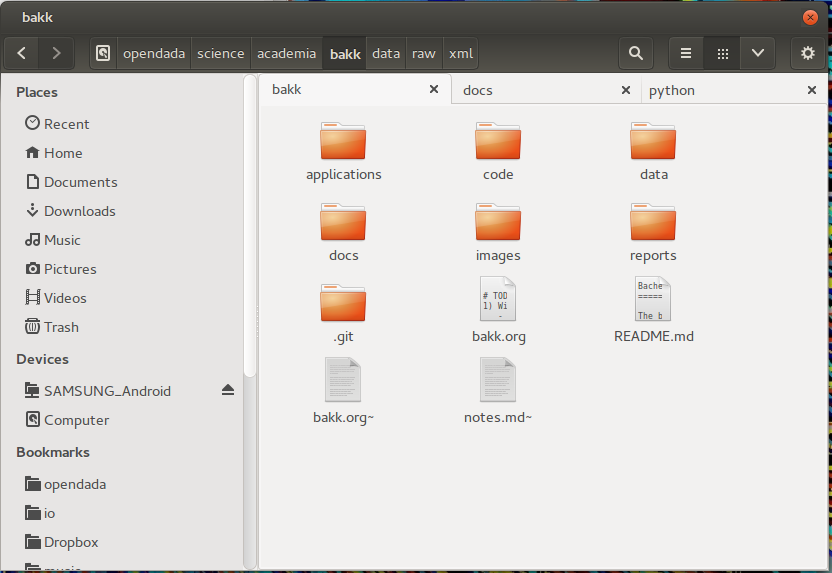
local folder structure
After creating the empty folder structure, I initialize a git-repo (git init) and add the README.md (markdown-template). In there I write all basic informations about the thesis, how to participate, license terms and the requirements necessary for it. Soon after this, the folders get filled with papers, some data and project files of software used – always seperated in raw and processed/edited by me. So, when the README.md is written, the LICENSE information is added and the basic structure created, I upload the first set of files onto my GitHub-repository. But beware of copyright issues: Control if you own the rights to share your content. If not gitignore helps you to not add files to your git repo.
openscienceASAP
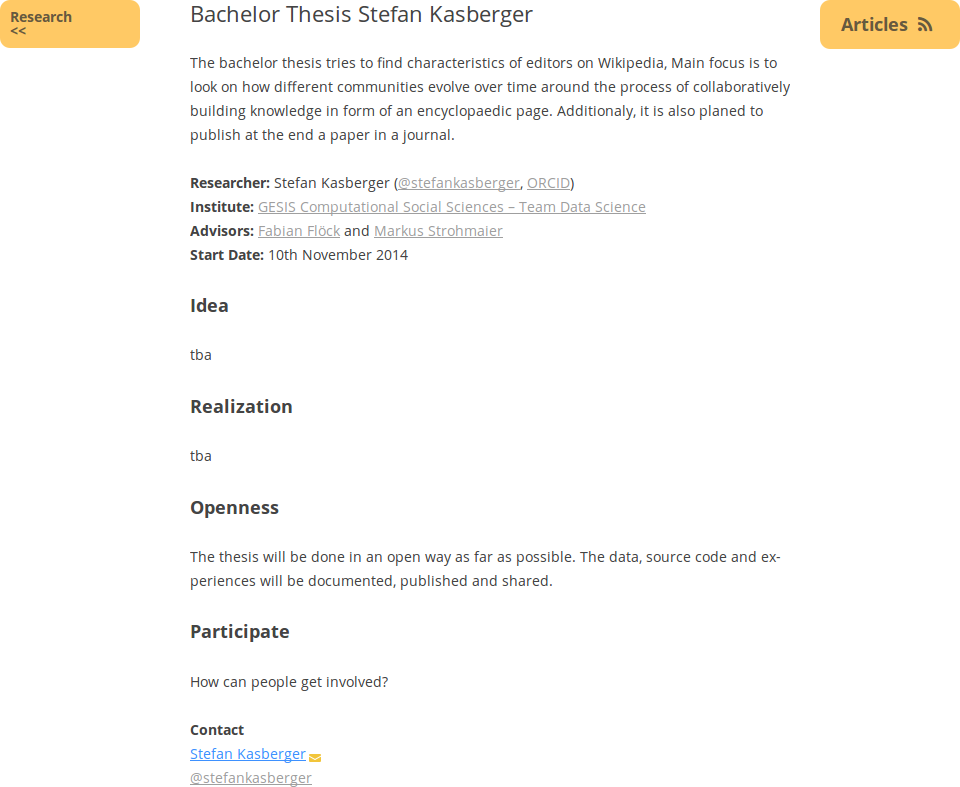
The last step is to add all information to an overview page here at openscienceASAP. This is the central point of contact for the scientific project and connects all dots, from blog posts over data and sourcecode to persons and their social media streams. Before adding the overview page (template), a new category as child of the research category will be created at the backend, so every post can be found via the category functionality (Category Bachelor Thesis Stefan Kasberger). Right now, it is still very empty, but it will be updated over time with the actual status.
Copyright
And finally: A crucial point is to think about copyright right from the beginning. For me, easy usage of my content is very important, but also that it is cited when it is reasonable. By default I use the MIT license for sourcecode, self-created data is under public domain and published text under CC BY 3.0 AT.